Thrust 1 — Imaging Biomarkers
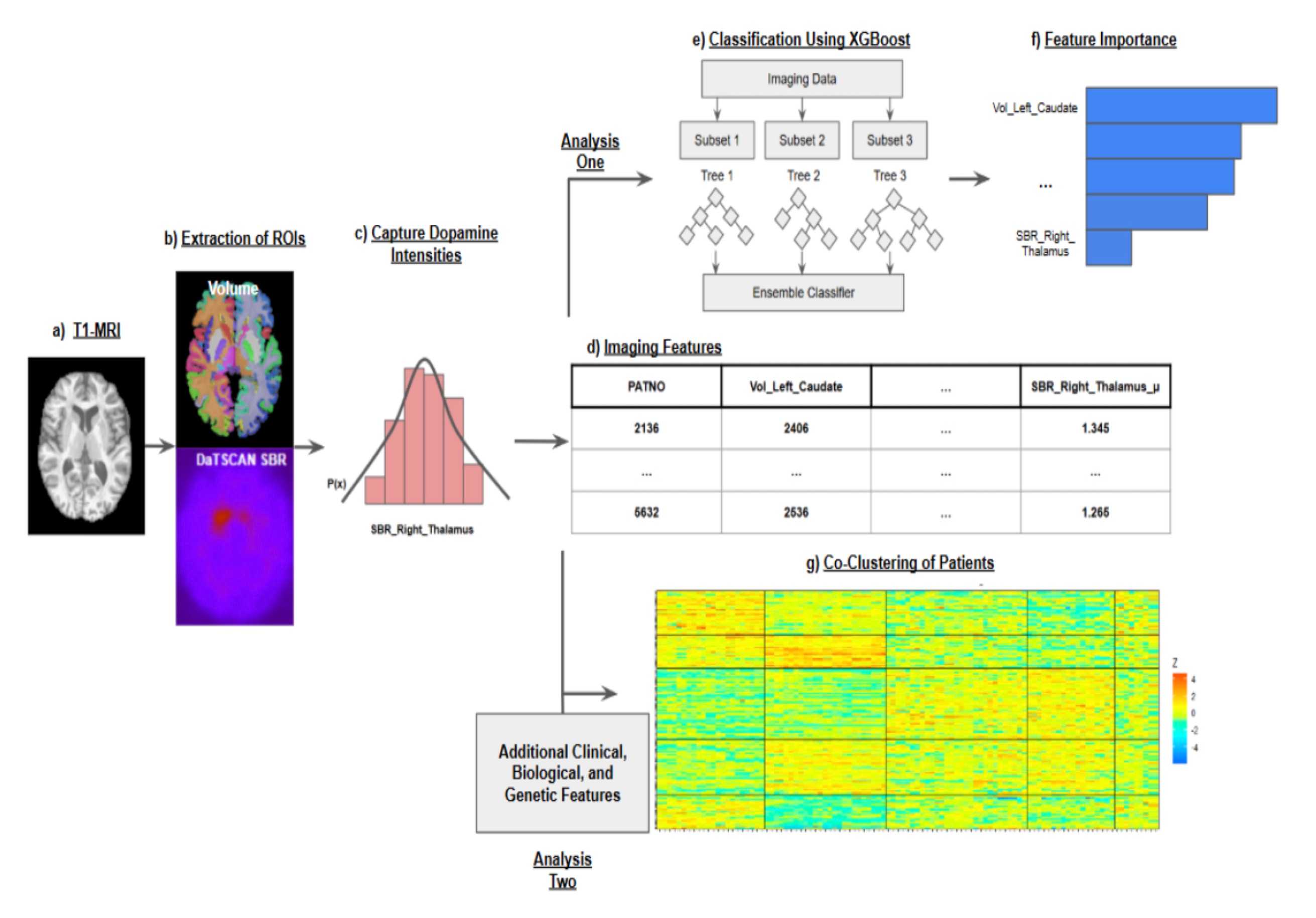
Automated DaT-SPECT and T1 MRI pipelines generate harmonized biomarker matrices that sharpen CSFSAA stratification and feed downstream agents.
We build actionable, multimodal agents that differentially diagnose Parkinson's mechanisms, surface subtype-specific biomarkers, and keep neurologists in the loop with interactive decision support.
Two coordinated pillars—domain thrusts and clinical workflows—move harmonized data from cohorts into bedside insight.
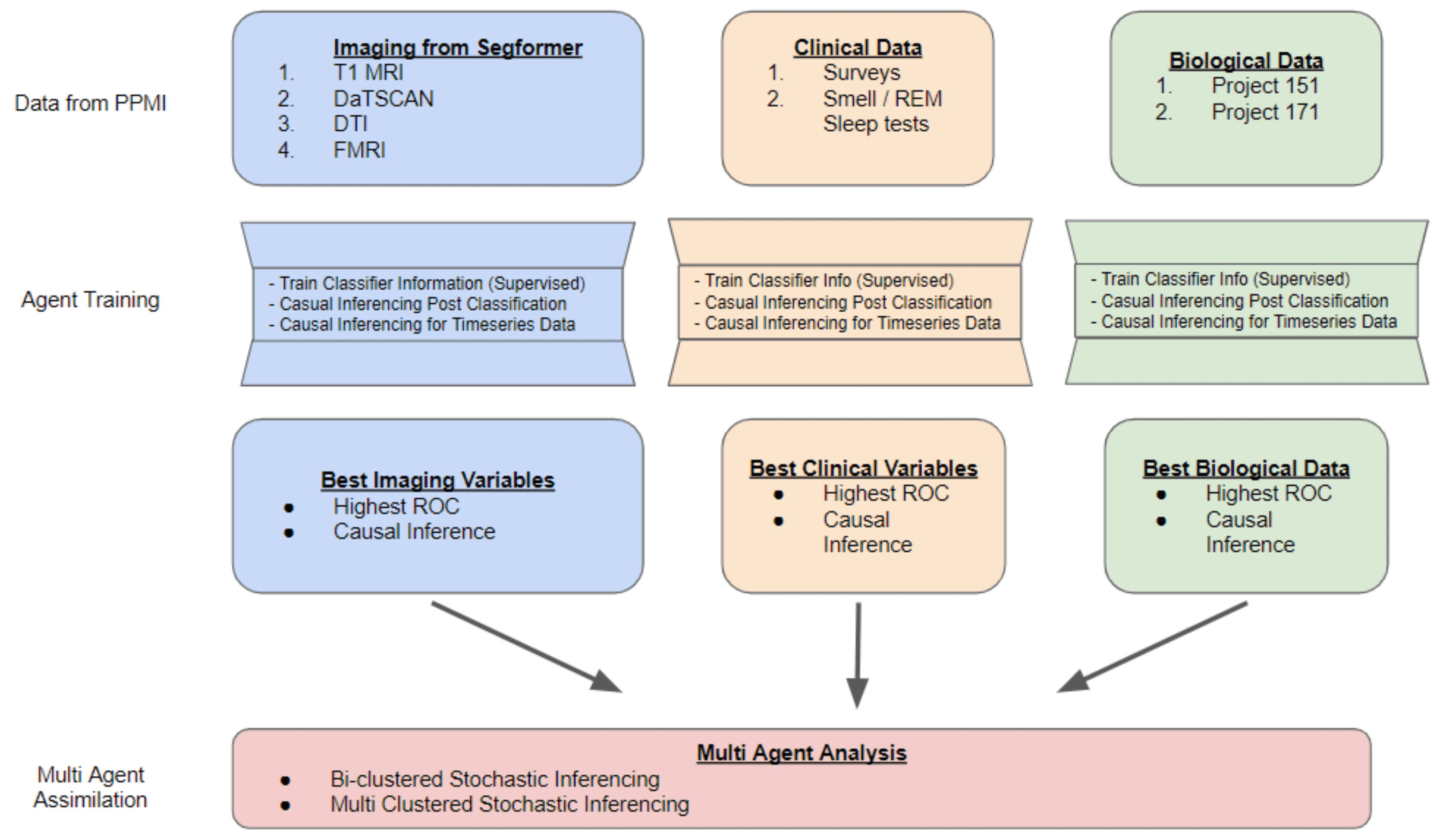
We harmonize PPMI and allied cohort assets—DaT-SPECT, T1 and diffusion MRI, biospecimens, genetics, gait sensors, and rich clinical batteries—into progressive agents that reason over time. Modality-specific thrusts extract stable features, while workflows coordinate generative modeling and clinician delivery.
The result: multimodal latent spaces that expose severity-aligned phenotypes, plus ActionIntel tooling that grounds recommendations in real-world practice.
Automated DaT-SPECT and T1 MRI pipelines generate harmonized biomarker matrices that sharpen CSFSAA stratification and feed downstream agents.
Free-water–corrected diffusion metrics align with clinical scales inside a compositional Bayesian co-clustering framework, revealing severity-aligned patient subtypes.
Wearable IMU data, cognitive batteries, and demographics combine to predict CSFSAA status and uncover multimodal phenotypes.
Large-scale statistical testing links multimodal biomarkers to putative biological pathways, grounding discoveries in mechanism-centric hypotheses.
See all thrusts on the Domain Thrusts page.
Harmonizes imaging, diffusion, and clinical data into interpretable latent spaces that seed state estimators and policy agents.
Delivers multimodal inferences to neurologists via interactive visualization, case review, and cohort monitoring tools.
Learn how modeling meets care on the Workflows page.
Strategic philanthropy, industry collaborations, and clinical trial partnerships accelerate progressive AI deployment. Visit the partners page to learn how funding and clinical collaborators engage, or connect through the resources portal.